A Name, mass and map of the circular minichromosome. The... Download Scientific Diagram

Welcome to RestrictionMapper - on line restriction mapping the easy way. Maps sites for restriction enzymes, a.k.a. restriction endonucleases, in DNA sequences. Also does virtual digestion.
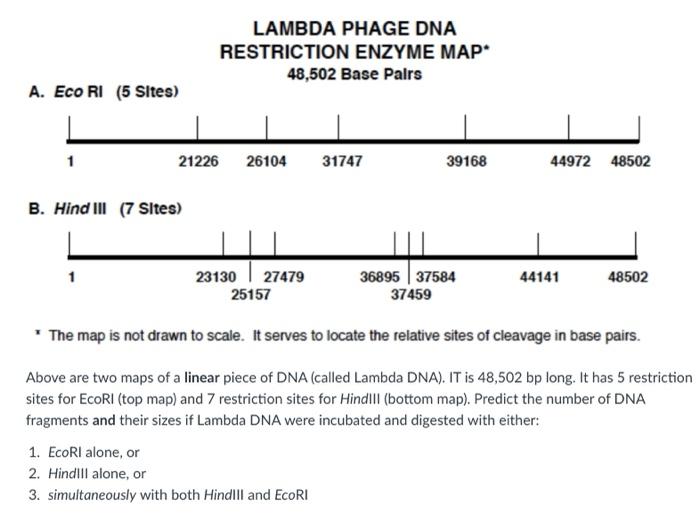
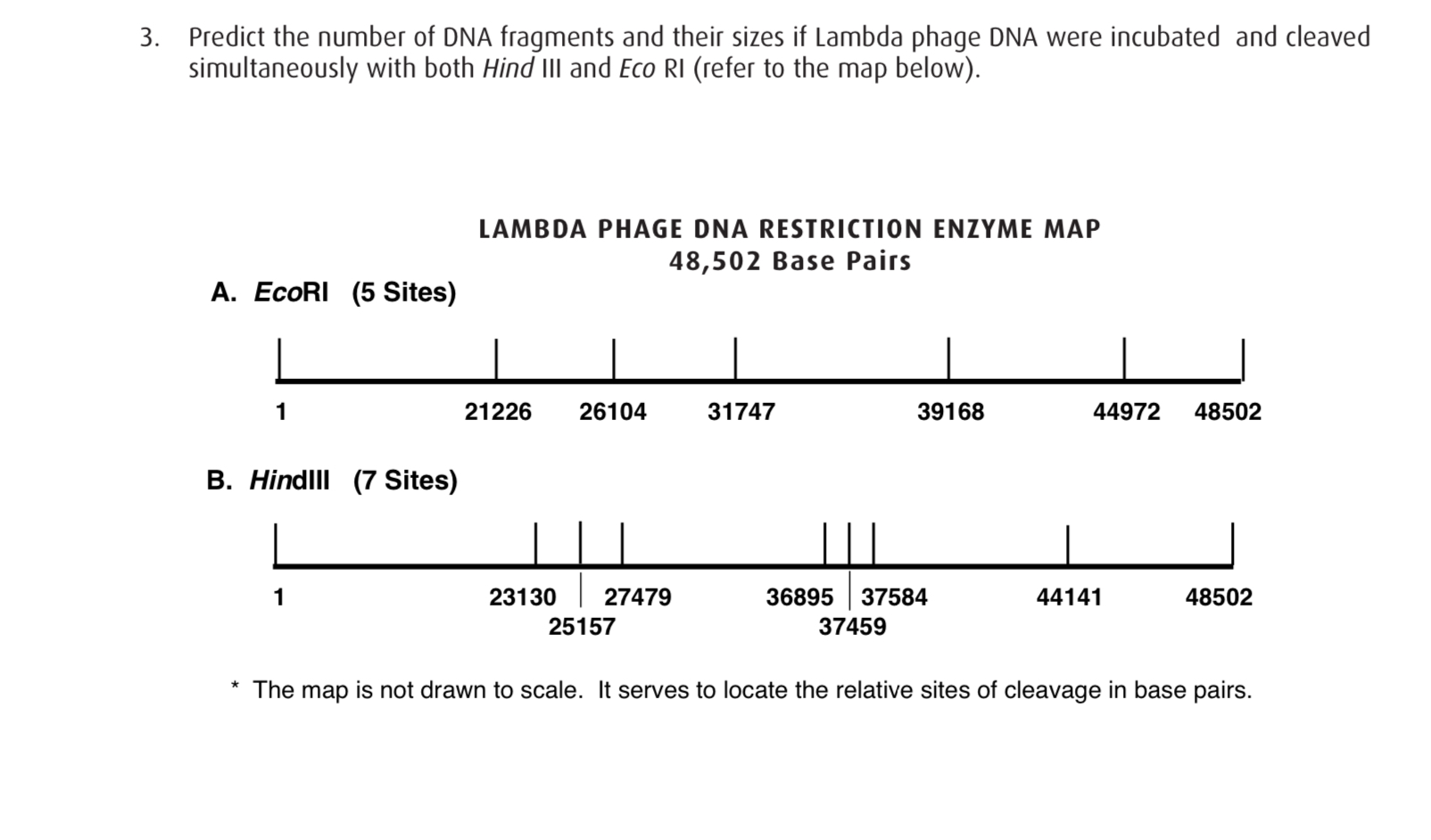
(Solved) LAMBDA PHAGE DNA RESTRICTION ENZYME MAP 48,502 Base Palrs A. Eco... (1 Answer
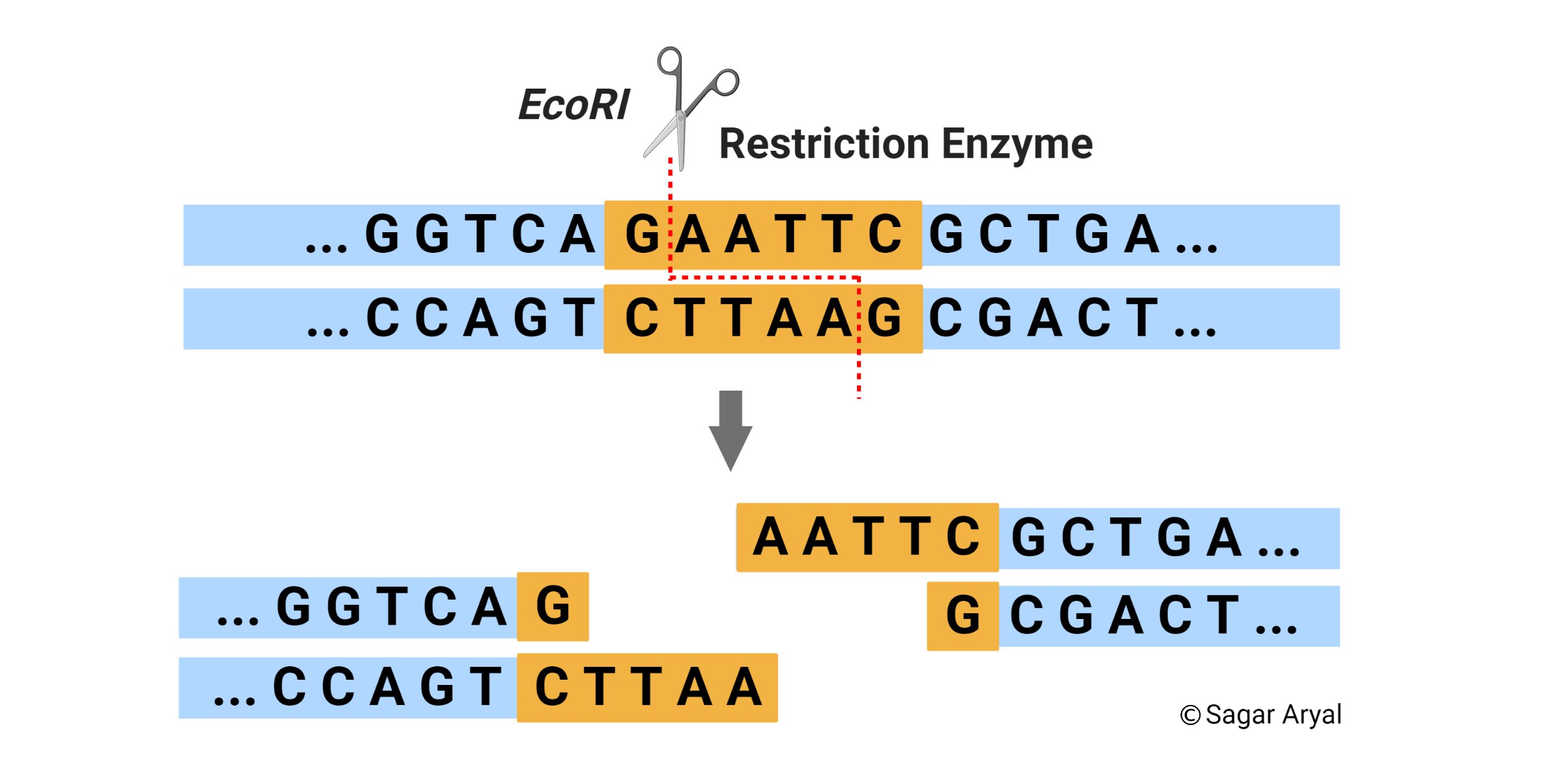
NEBcutter2 is a tool that allows you to analyze DNA sequences for restriction enzyme sites and perform virtual digestion. You can also compare your results with other NEB tools, such as Enzyme Finder and NEBioCalculator, and access the NEB website for more information on molecular biology products and services.
Which of the Following Describe Restriciton Enzymes and Their Use GriffinkruwHowell
A map of bacteriophage lambda was constructed, including accurate positions for all 41 cut sites made by 12 different restriction enzymes. Over 100 fragments from single, multiple, and partial enzyme digestions were measured versus standards that were calibrated with respect to DNA molecules of know.
Restriction Enzyme Map

From that list select Lambda. This is the name of the DNA you will be digesting. Select the Linear option where the tool says "the sequence is" and the "NEB enzymes" option where the tool says, "Enzymes to use". Then click Submit. The next page will display a full restriction map of the lambda DNA.
Restriction mapping of circular DNA YouTube
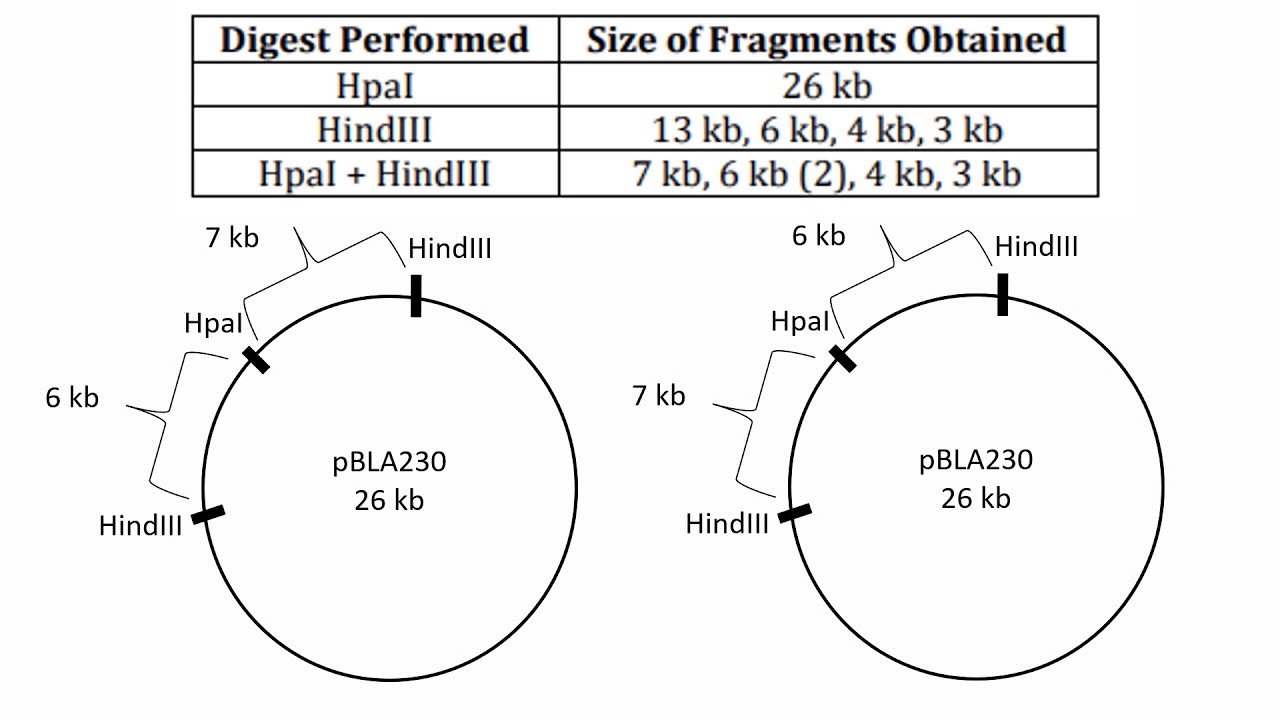
unknown DNA fragments. Restriction enzymes were a catalyst for the molecular biology revolution, and now hundreds of such enzymes are known. In this investigation, the restriction enzymes EcoRI, PstI, and HindIII will be used to digest bacteriophage lambda DNA. Gel electrophoresis will be employed to separate the resulting DNA fragments,
PPT Restriction Analysis of Plasmid DNA PowerPoint Presentation, free download ID2351797

In the cleavage reaction bytwo restriction enzymes, choose the reaction enzyme buffer with the higher ionic strength. Prepare a sample ofuncleaved DNA for the gel bydiluting 5/21 of lambda DNA with 45/al ofsterile wat Swirl r.the tubes gently andincubate at 37°C. After onehour, add 5/21 Stop Buffer toeach reaction vessel and heat to65°C for.
Restriction map of cosTUB2 and functional analysis. Linear... Download Scientific Diagram

1, 2 and 3. Into tube No 1, place 8 p~l of EcoRI-digested lambda DNA, into tube No 2 place 8 p.l of HindIII-digested lambda DNA, and into tube No 3 place 8 p.l of lambda DNA digested with both of these restriction enzymes. Heat the tubes for 5 min in a 65°C water bath. This insures
Plasmid Restriction Enzyme Maps

For cleavage with two restriction enzymes, combine 34 M1 sterile water, 5 pd of 10 X restriction enzyme buffer, 3 p1 of each restriction enzyme and 5 pl of lambda DNA. In the cleavage reaction by two restriction enzymes, choose the reaction enzyme buffer with the higher ionic strength. Prepare a sample of uncleaved DNA for the gel by diluting 5.
lambda Sequence and Map
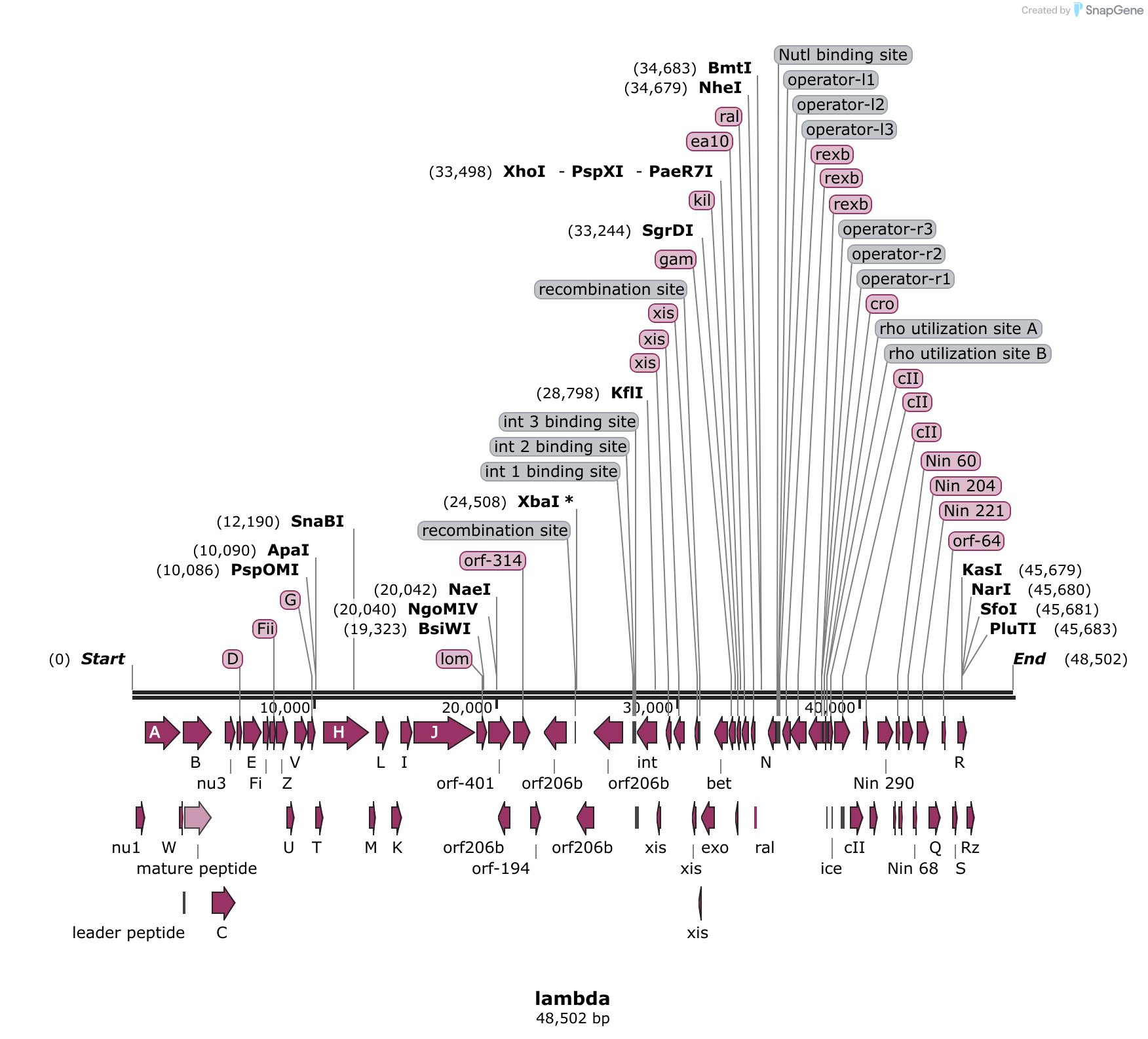
Ron Davis at Stanford developed phage lambda into a powerful cloning vector by showing that some restriction enzymes leave sticky ends and then utilizing these enzymes for DNA cloning (Mertz and Davis, 1972; Cameron et al., 1975), Kaiser and Masuda (1973) devised a procedure for packaging exogenously added lambda DNA into lambda virions, and.
Restriction maps of plasmids used in this study. The sites are B1,... Download Scientific Diagram

Site-specific λ-DNA modification strategies generally fall within one of four categories: (i) restriction enzyme cleavage and ligation 4, 8; (ii) recombinase-mediated modifications 9, 10; and.
Answered 3. Predict the number of DNA fragments… bartleby
Abstract. A map of bacteriophage lambda was constructed, including accurate positions for all 41 cut sites made by 12 different restriction enzymes. Over 100 fragments from single, multiple, and partial enzyme digestions were measured versus standards that were calibrated with respect to DNA molecules of known sequence.
Vector Plasmid Restriction Map Restriction Enzyme Multiple Cloning Site, PNG, 687x600px, Vector
This is a BamHI-EcoRI restriction map of Lambda DNA: (read it like this: (e.g.) "the 4878 bp long EcoRI fragment is between the first and the second EcoRI site, and in the Simple Cloning Lab it is called Eco E" ).. An output file from EMBOSS>restrict for restriction enzyme sites in Lambda DNA can be found here. The list shows all enzymes.
Example DNA sequence and its optical map corresponding to XhoI... Download Scientific Diagram

Perform a digestion of lambda DNA with HIndIII, EcoRI, and PstI restriction enzymes. Load and run digested DNA through Electrophoresis. Generate a standard curve based on lambda HindIII DNA Standard. Determine length of all DNA fragments by comparing to the standard curve.
9. On Worksheet 16.IIIB is a restriction map of bacteriophage lambda. You digest some lambda DNA

The sizes were determined by comparison to a molecular ladder which has bands of known sizes when it is separated by electrophoresis at the same time as the digested λ DNA. Restriction sites of Lambda (λ) DNA - in base pairs (bp) The sites at which each of the 3 different enzymes will cut lambda DNA are shown in the maps Enzymes A, B and C below.
Addgene SLBP nontargeting gRNA lambda phage

A restriction enzyme map is a basic requirement for the detailed. (Hind III digest of phage lambda DNA) from any reputable molecular biology company (store at -20°C); iso-. Restriction of pXl108 DNA Set up the restriction enzyme digestions shown in Table 1, using 0.5 ml microcentrifuge tubes.
PPT RESTRICTION DIGESTION AND ANALYSIS OF LAMBDA DNA PowerPoint Presentation ID1606247

The map below shows the positions of all known ORFs larger than 200 codons. Enzymes with unique restriction sites are shown in bold type and enzymes with two restriction sites are shown in regular type. Coordinates indicate position of cutsite on the top strand. In previous catalogs, coordinates referred to the